Darren J. Lighter, Clyde Pruett, Cody Wolf, Joseph Tuccinardi, Riley Olsen, Dr. Matthew D. King, Dr. Ken Tawara, Dr. Don L. Warner, Dr. Cheryl L Jorcyk
Abstract
Approximately 1 in 8 women in the United States will develop breast cancer in their lifetime. While localized breast cancer has a 5-year survival rate of 99%, metastatic breast cancer has a limited 5-year survival rate of 27%. Prior research from our lab and others have identified a proinflammatory cytokine that induces invasion and detachment of various breast cancer tumor cells. We hypothesize that small molecule inhibitors (SMI), can bind to proinflammatory cytokines and reduce their potential to activate the signaling pathway that promotes metastasis of breast cancer tumor cells. First, an in silico model was used to identify potential parental inhibitors based on prospective binding affinity. Parental molecules were then synthesized through a multi-step organic synthesis, and optimized analogs were presented for potentially increased binding affinity and drug-like properties. Continuing forwards, various in vitro tests were performed, including enzyme linked immunosorbent assay (ELISA) and western blots, which allow us to compare the effectiveness of each SMI to reduce the signaling cascade. Inhibition of these cytokines may lead to novel therapeutics that can be used to prevent breast cancer metastasis.
Introduction
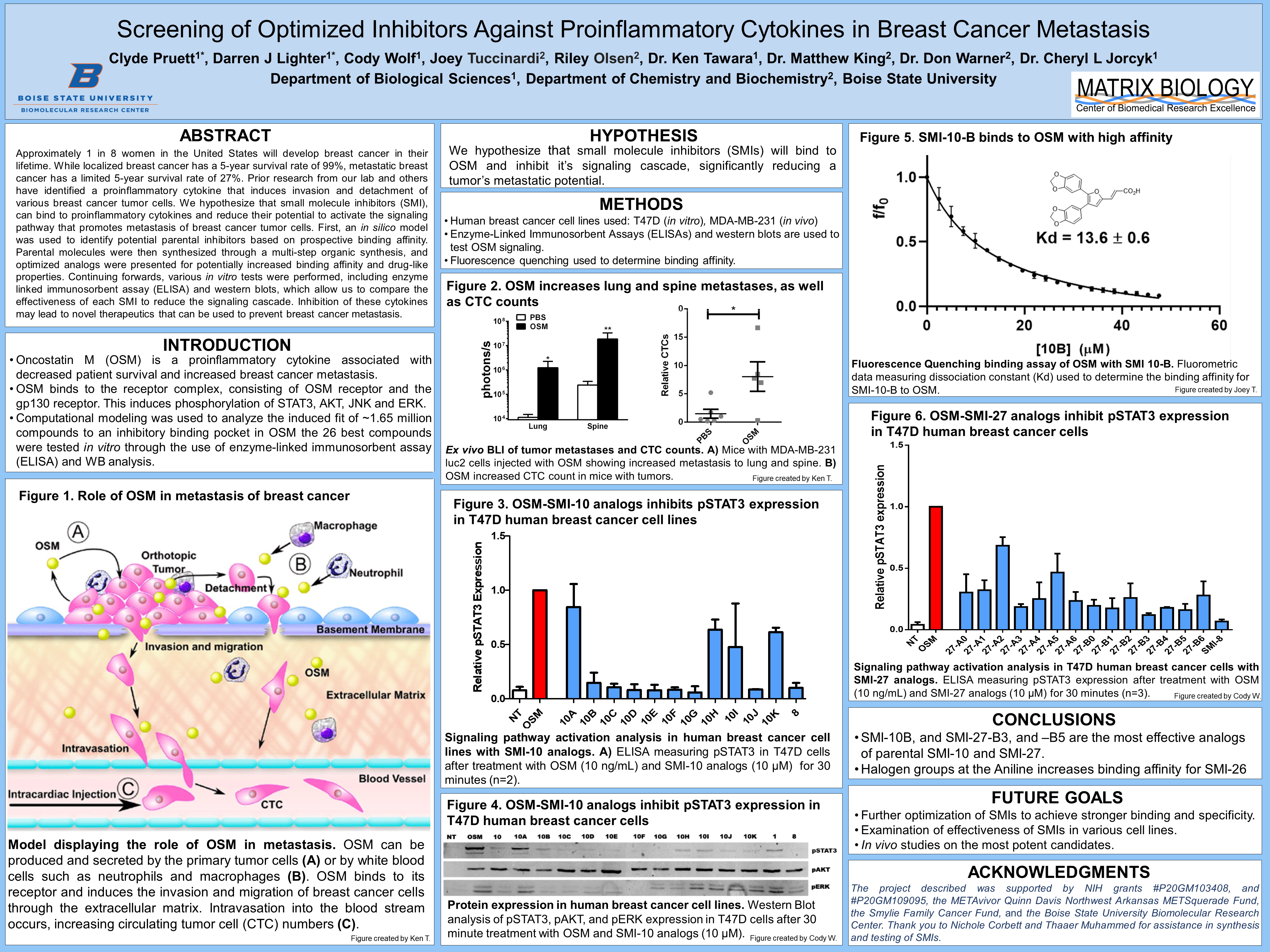
- Oncostatin M (OSM) is a proinflammatory cytokine associated with decreased patient survival and increased breast cancer metastasis.
- OSM binds to the receptor complex, consisting of OSM receptor and the gp130 receptor. This induces phosphorylation of STAT3, AKT, JNK and ERK.
- Computational modeling was used to analyze the induced fit of ~1.65 million compounds to an inhibitory binding pocket in OSM the 26 best compounds were tested in vitro through the use of enzyme-linked immunosorbent assay (ELISA) and WB analysis.
Figure 1. Role of OSM in metastasis of breast cancer Model displaying the role of OSM in metastasis
Hypothesis
We hypothesize that small molecule inhibitors (SMIs) will bind to OSM and inhibit it’s signaling cascade, significantly reducing a tumor’s metastatic potential.
Methods
- Human breast cancer cell lines used: T47D (in vitro), MDA-MB-231 (in vivo)
- Enzyme-Linked Immunosorbent Assays (ELISAs) and western blots are used to test OSM signaling.
- Fluorescence quenching used to determine binding affinity.
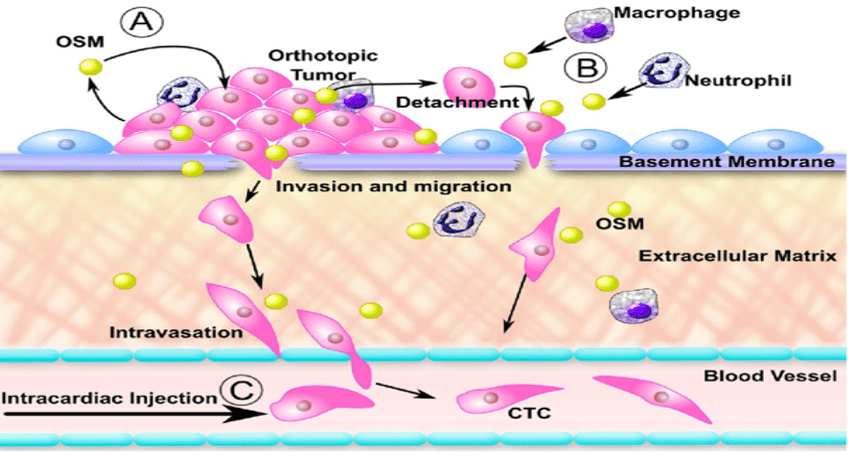
Figure 2. OSM increases lung and spine metastases, as well as CTC counts
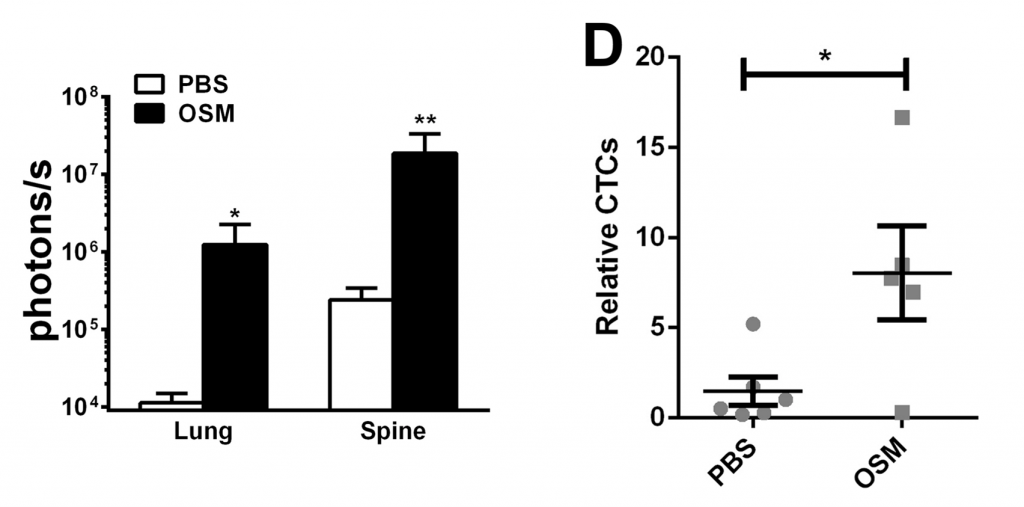
Figure 3. OSM-SMI-10 analogs inhibits pSTAT3 expression in T47D human breast cancer cell lines
Figure 4. OSM-SMI-10 analogs inhibit pSTAT3 expression in T47D human breast cancer cells
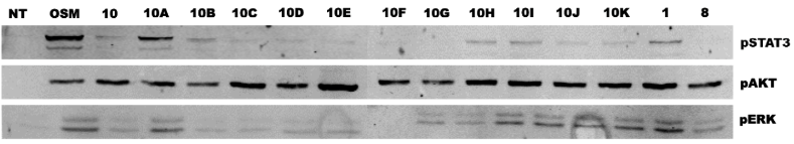
Figure 5. SMI-10-B binds to OSM with high affinity
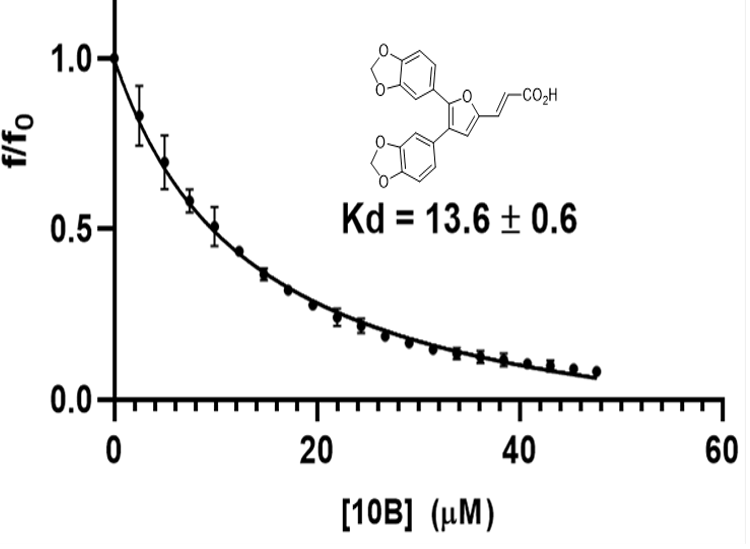
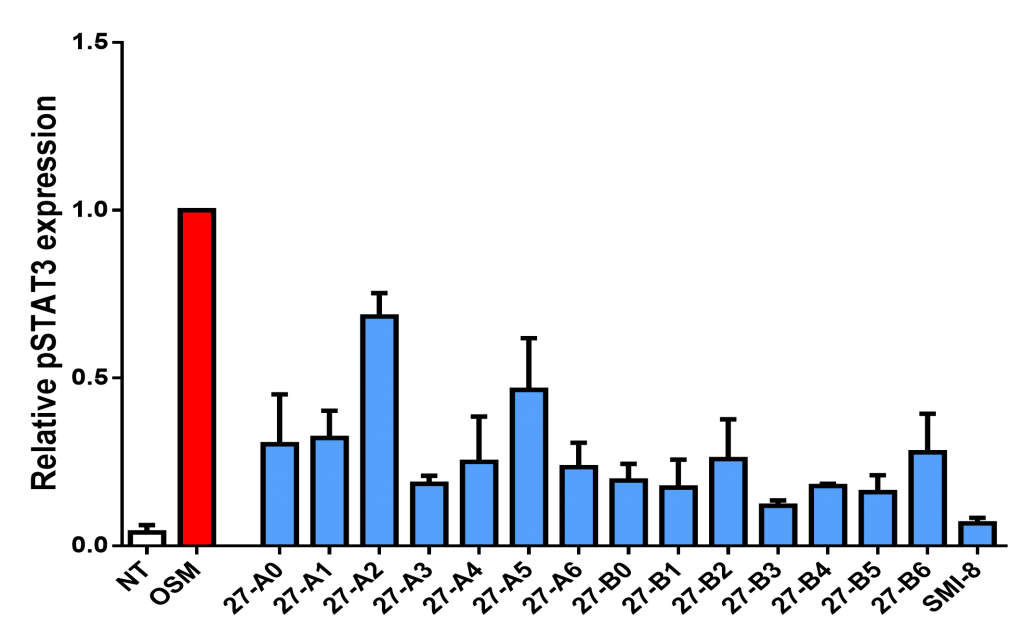
Figure 6. OSM-SMI-27 analogs inhibit pSTAT3 expression in T47D human breast cancer cells
Conclusions
- SMI-10B, and SMI-27-B3, and –B5 are the most effective analogs of parental SMI-10 and SMI-27.
- Halogen groups at the Aniline increases binding affinity for SMI-26
Future Goals
- Further optimization of SMIs to achieve stronger binding and specificity.
- Examination of effectiveness of SMIs in various cell lines.
- In vivo studies on the most potent candidates.
Acknowledgements
The project described was supported by NIH grants #P20GM103408, and #P20GM109095, the METAvivor Quinn Davis Northwest Arkansas METSquerade Fund, the Smylie Family Cancer Fund, and the Boise State University Biomolecular Research Center. Thank you to Nichole Corbett and Thaaer Muhammed for assistance in synthesis and testing of SMIs.
Additional Information
For questions or comments about this research, contact Darren Lighter at darrenlighter@u.boisestate.edu.