Joey Day, Kyle Fisch, Joseph Tuccinardi, Riley Olsen, Dr. Don L. Warner
Abstract
We are interested in an inflammatory cytokine (IC) that is overexpressed in metastatic breast cancer, which is significant because there are more than 200,000 new cases of breast cancer diagnosed per year and it is expected to cause 42,690 deaths in 2020. The same IC is present in other inflammatory diseases such as systemic scleroderma, irritable bowel disease, and cystic fibrosis. The IC acts upon the JAK-STAT pathway, which causes gene transcription of inflammatory factors. Monoclonal antibodies which bind to the IC are in clinical trials but are an expensive solution. In order to find a more accessible treatment, small molecule inhibitors were screened computationally to bind to the IC. SMI-10 and SMI-27 showed successful binding through ELISA so we are using those as a base to synthesize novel compounds that bind with high affinity to the IC. By continually producing unique SMIs, we are simultaneously synthesizing potential drug candidates for treatment of diseases related to the IC as well as informing future SMI design. Ultimately, these binding studies will inform our drug design efforts to develop a cheaper, easily administered treatment for diseases caused by overexpression of the IC.
Background: IC Inflammation Pathway and SMI Structures

Previous Work: Select Analogs of Lead Compounds

B) Pyrimidine amide based SMI
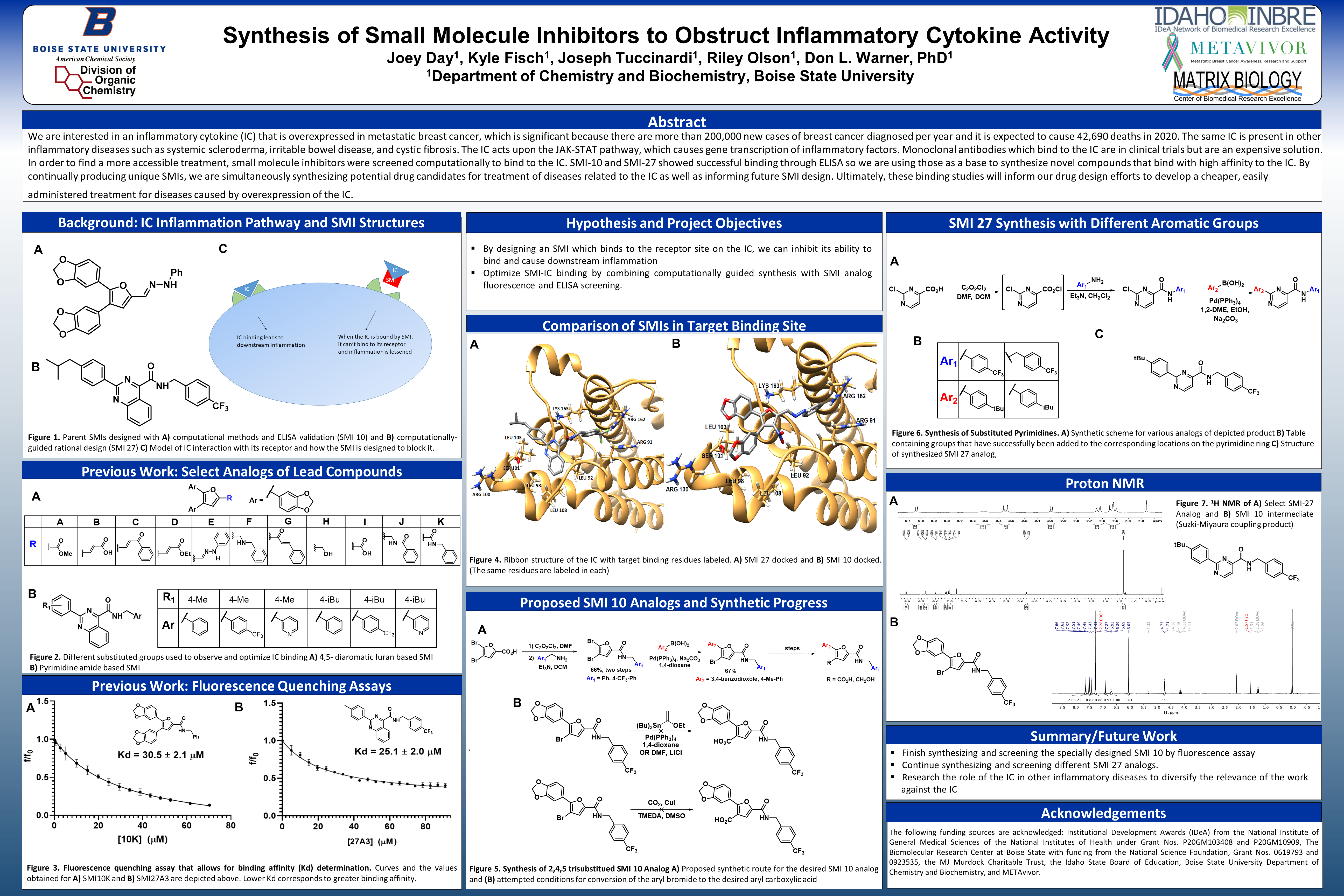
Previous Work: Fluorescence Quenching Assays
Hypothesis and Project Objectives
- By designing an SMI which binds to the receptor site on the IC, we can inhibit its ability to bind and cause downstream inflammation
- Optimize SMI-IC binding by combining computationally guided synthesis with SMI analog fluorescence and ELISA screening.
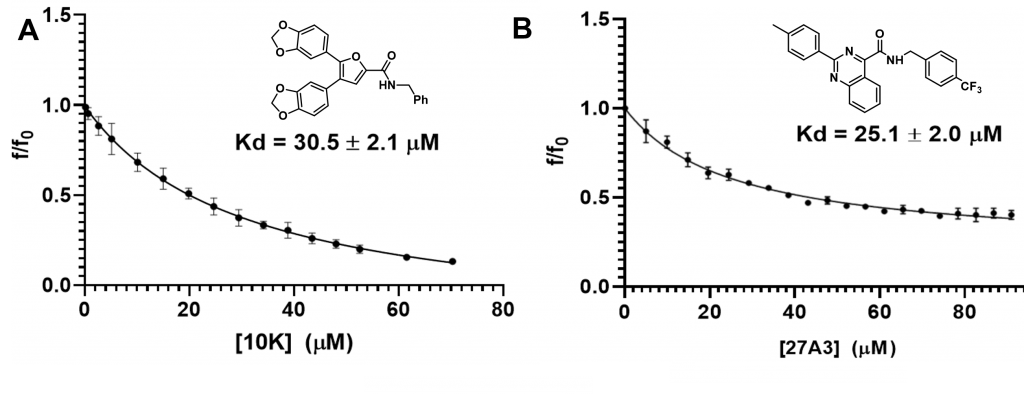
Comparisons of SMIs in Target Binding Site
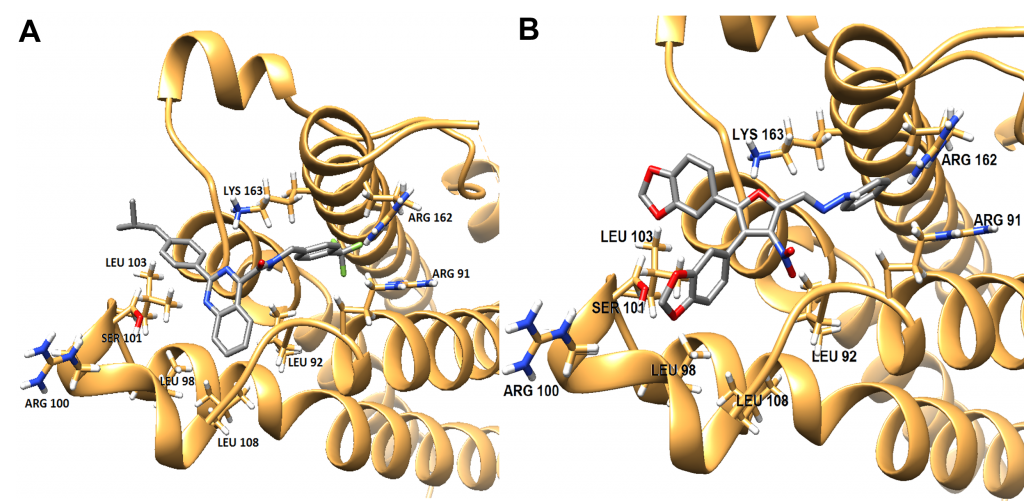
Proposed SMI 10 Analogs and Synthetic Progress
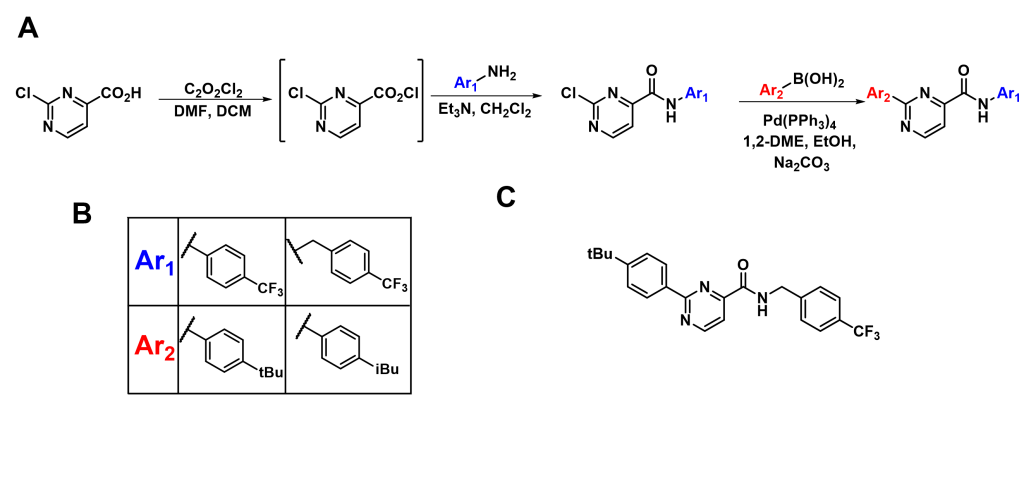
SMI 27 Synthesis with Different Aromatic Groups
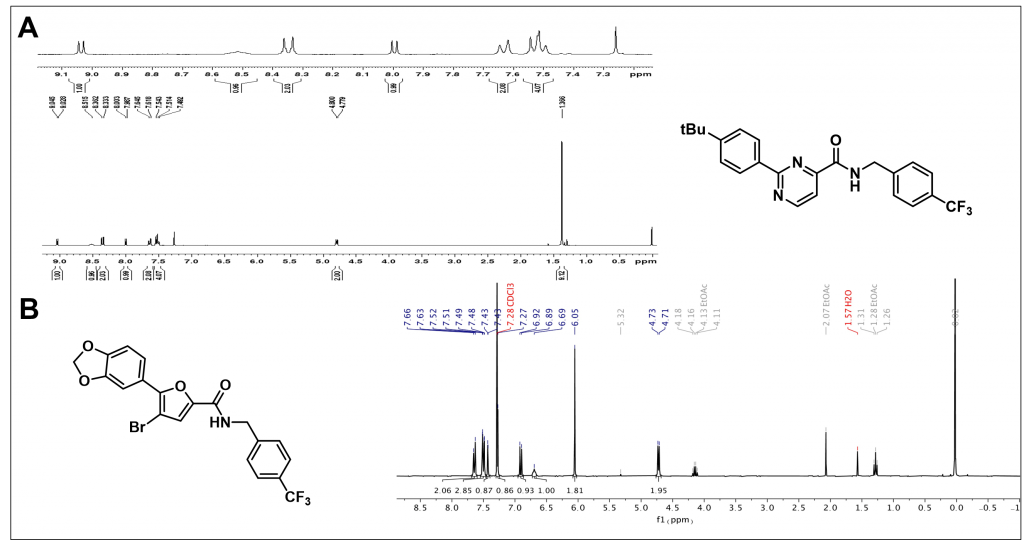
Proton NMR
Summary/Future Work
- Finish synthesizing and screening the specially designed SMI 10 by fluorescence assay
- Continue synthesizing and screening different SMI 27 analogs.
- Research the role of the IC in other inflammatory diseases to diversify the relevance of the work against the IC
Acknowledgements
The following funding sources are acknowledged: Institutional Development Awards (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under Grant Nos. P20GM103408 and P20GM10909, The Biomolecular Research Center at Boise State with funding from the National Science Foundation, Grant Nos. 0619793 and 0923535, the MJ Murdock Charitable Trust, the Idaho State Board of Education, Boise State University Department of Chemistry and Biochemistry, and METAvivor.
Additional Information
For questions or comments about this research, contact Kyle Fisch at kylefisch@u.boisestate.edu.