About
Associate Professor
Biochemistry
Email: lisawarner@boisestate.edu
Office: ERB 3125
Phone: (208) 426-2260
Research Interests:
- Structural Biochemistry
- Protein Chemistry
- Drug discovery
- Spectroscopy
Research and Publications
Undergraduate Research Assistant
What courses would you recommend students have taken prior to working in your laboratory?
General Chemistry, ideally Organic and Analytical, but you will be considered even if you have not. Sign up for VIP 200 if you are in Freshman or Sophomore year.
How many hours per week do you expect a student to spend in the laboratory per research credit and does the time have to be a set schedule?
10 hours per week is pretty typical. When you are new to the lab, you will need to adhere to a set schedule.
Ideally, for what length of time would a student research in your laboratory to achieve a meaningful research experience?
1 year including a summer internship provides adequate time to develop research skills and to engage in a meaningful research experience.
Educational Background
2011: Ph.D. – Biochemistry | University of Colorado
2002: B.S. – Chemistry | Boise State University
Research Overview

Protein and RNA interactions
We use integrated structural biology to tackle questions about the structure and function of proteins, RNAs, and complexes in solution.
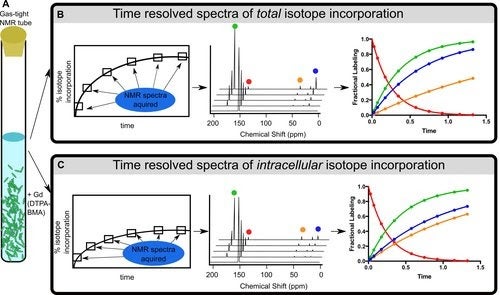
In-cell NMR
Our goal is to develop in-cell NMR techniques in order to build metabolic flux models of carbon utilization in microbes that are important in biofuels/chemicals production. In collaboration with scientists at the National Renewable Energy Laboratory (NREL), we use NMR to detect the conversion of single atom isotope labeled metabolite tracers in living cells.
Structure-based drug design
We work together on teams of researchers aiming to develop next-generation drugs for the treatment of diseases including cancer and heart disease.
Select Publications (2018-2021)
Mass, O., Tuccinardi, J., Woodbury, L., Wolf, C., Grantham, Bri., Pu, X., King, M.D., Warner, D.L., Jorcyk, C., Warner, L.R., “Bioactive Recombinant Human Oncostatin M for NMR-based Screening in Drug Discovery.” Sci. Rep. (in revision) (2021)
Cantrell, M.S., Soto-Avellaned, A., Wall, J.D., Ajeti, A.D., Morrison, B.E., Warner, L.R., McDougal, O.E., “Repurposing drugs to treat heart and brain illness.” Pharmaceuticals in press. (2021)
Oxford, J. T., … Warner, L.R. et al. “Center of Biomedical Research Excellence in Matrix Biology: Building Research Infrastructure, Supporting Young Researchers, and Fostering Collaboration.” Int. J. Mol. Sci. 21, 2141 (2020).
Marquart, L.A., Turner, M.W., Warner, L.R., King, M.D., Groome, J.R., & McDougal, O.M. (2019). “Ribbon α-Conotoxin KTM Exhibits Potent Inhibition of Nicotinic Acetylcholine Receptors.” Marine Drugs, 17 (12) 669.
Warner, L.R., Mass, O., Lenn, N.D., Grantham, B.R., Oxford, J.T. (2018) “Growing and Handling of Bacterial Cultures within a Shared Core Facility for Integrated Structural Biology Program”, IntechOpen, DOI: 10.5772/intechopen.81932.
Pabis, M., Popowicz, G.M., Stehle, R., Fernández-Ramos, D., Asami, S., Warner, L.R., García-Mauriño, S.M., Schlundt, A., Martínez-Chantar, M.L., Díaz-Moreno, I., Sattler, M. (2018) “HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs”, Nucleic Acids Research, 47, 1011–1029. DOI: https://doi.org/10.1093/nar/gky1138
Select Grants (2018-2021)
7/1/2019 - 5/1/2022
Center of Biomedical Research Excellence in Matrix Biology Phase II – National Institutes of Health
8/1/2019 – 7/31/2022
Mechanistic Investigation on Carrier Protein Recognition in Quorum Sensing Synthases – National Science Foundation
6/1/2020 – 5/31/2022
Using Fortilin Inhibitors to Halt Atherosclerosis Year 2020 – University of Washington/NIH pass thru
5/1/2021 – 4/30/2024
Regulation of Collagen Type I Expression by Chaperone-Mediated mRNA Remodeling – National Institutes of Health / DHHD
10/1/2019 – 9/30/2022
Molecular Mechanisms of the Posttranscriptional Regulatro LARP6 – Texas State University
9/15/2017 – 8/31/2021
RII Track-4: using in cell NMR to follow 13C-fluxomics in Living Cells – National Science Foundation