The Nagarajan Lab has an opening for a Postdoctoral Fellow. Please see the Job Openings section on his Lab’s website for details regarding this position.
Contact Information
Professor
Biochemistry
Email: rajnagarajan@boisestate.edu
Office: SCNC 313
Phone: (208) 426-1423
Research Interests
- Antibiotic Resistance, Bacterial Virulence and Quorum Sensing
- Enzymology / Antimicrobial Drug Discovery
- Bioorganic Chemistry / Protein Biochemistry / Chemical Biology
- Biomolecular NMR to investigate structure-function relationships in proteins
Educational Background and Research
Educational Background
- 2004-2006: MD Postdoc | Johns Hopkins University, Baltimore
- 1998-2004: Ph.D. – Chemistry | Wesleyan University, Middletown, CT
- 1996-1998: M.S. – Chemistry | Indian Institute of Technology, Chennai
- 1993-1996: B.S. – Chemistry | Madras Christian College, Chennai, India
Research Focus



In the Nagarajan laboratory, we investigate AHL synthase enzymes (I-proteins) to develop quorum sensing inhibitors. Students working in these projects will have the opportunity to learn a variety of tools/skills such as small molecule synthesis, protein expression and purification, protein biochemistry and chemical biology tools, and enzyme kinetics. Depending on the research project, students receive hands-on experience on a broad range of biophysical / analytical instrumentation such as FPLC, HPLC, NMR, Steady State fluorescence, UV-Visible spectrophotometry, colorimetry, stopped flow fluorescence (fast kinetics) etc. We collaborate with structural biologists (X-ray crystallography, biomolecular NMR spectroscopy, Molecular Dynamic simulations etc) to address mechanistic questions in quorum signal synthesis. Students working in these projects also get exposure to bioinformatic tools, molecular visualization tools (Chimera, PyMol etc.) protein homology modeling, and molecular docking tools.
Research Overview
The rapid rise in multidrug resistant organisms pose special challenges to treating bacterial infections. Therefore, therapeutic strategies that combat bacterial virulence without aggravating drug resistance are in great demand. Since antibiotics threaten the survival of microorganism, it puts selective pressure on bacteria to develop resistance. Therefore, compounds that doesn’t directly kill bacteria (antibiotics) but rather limits the microbe’s ability to harm the host (antivirulence) are attractive as novel antimicrobials in antibacterial therapy.
When and how does bacteria turn virulent?
A single cell bacterium in the planktonic mode do not have the means to impact the environment. In this state, bacteria are typically avirulent. In most instances however, bacteria seldom remain in the planktonic mode but instead communicate and cooperate with their immediate neighbors to behave like a multicellular species. In the ‘social/group mode’ bacteria makes collective decisions to fight off common threats such as a looming antibiotic attack, immune response from the host during infection, responding to resource strains in the environment etc. Interbacterial communication helps bacteria transition from an avirulent planktonic state to a virulent social mode. This communication is facilitated by a chemical signal sensing mechanism called “Quorum Sensing”.
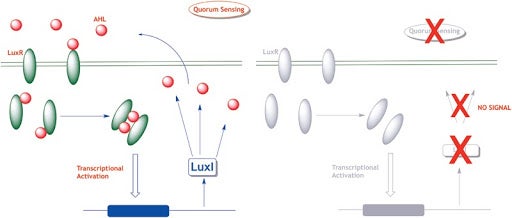
How does Quorum Sensing work?
Bacteria makes specific signal molecules called autoinducers. While Gram-positive bacteria use peptide signaling, Gram-negative bacteria predominantly use Acyl Homoserine Lactones (AHLs) chemical signals to facilitate quorum sensing. A dedicated set of proteins called the I-proteins or AHL synthases make these intracellular AHL signals that are eventually released to the local environment. Since signal strength in the environment reflect local population-levels, bacteria count these signals to estimate its local population density and hence the name quorum sensing (QS). QS informs bacteria if their local population has reached quorum levels necessary to trigger social traits such as forming a biofilm, releasing virulence factors, infecting a host etc. Since a larger population gives competitive advantage for bacteria to survive immune / antibiotic onslaught, this information is critical for bacteria to judge the optimum timing to infect a host. Quorum sensing inhibitors would interfere with bacteria’s ability to countits population and keep them in an avirulent mode. The long-term focus in our lab is to develop quorum sensing modulators that either inhibit or activate QS in bacteria (can you think of a reason on how QS activators could also become useful tools to fight bacterial infections?). In addition to their biomedical significance, QS modulations may also serve as useful mechanistic probes to study social behavior in bacteria (sociomicrobiology).
Current Projects
Medicinal Chemistry/Drug Discovery
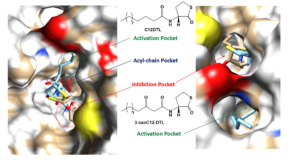
In this project, we use a medicinal chemistry approach to synthesize a library of AHL QS signal analogs and use them as chemical probes to uncover novel binding pockets in AHL synthases. Student training opportunities in this project include small molecule synthesis, protein purification, enzyme inhibition assays and mechanism of inhibition studies. The outcome of this project will reveal a) new pharmacophores for developing antivirulence-based medicinal compounds and b) mechanistic probes for studying social behavior in bacteria.
Molecular Biophysics
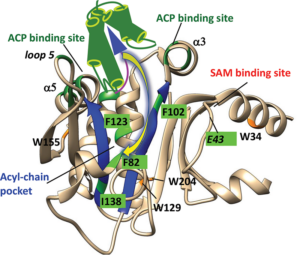
The goal of this project is to develop chemical tools for determining cargo-flipping rates in ACP dependent enzyme. We use a stopped-flow fluorometer to measure pre-steady state kinetics (fast kinetics) in the milliseconds to seconds time range of substrate binding to enzymes. Students working in this project will learn to use genetic code expansion tools to incorporate fluorophores at specific sites in proteins. The tools discovered in this project will reveal the mechanism of carrier protein recognition with their partner enzymes.
Mechanistic Enzymology
Fidelity / Specificity in signal synthesis is an important aspect of quorum sensing, yet the molecular details on how AHL synthases achieve fidelity in signal synthesis continue to remain poorly resolved. This project involve mechanistic studies on a library of AHL synthases using the tools of protein biochemistry, enzymology, chemoenzymatic synthesis and molecular modeling.
Protein NMR
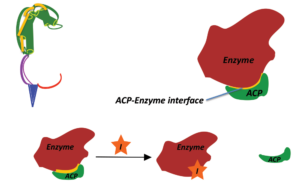
In this project, we use biomolecular NMR tools (collaboration) to map protein-protein contact surfaces in AHL signal synthesis. Results from this project will enhance our understanding of substrate recognition in QS signal synthesis and open new doors for developing small-molecules that interrupt protein-protein interface in QS signal synthesis.
Structural Biology
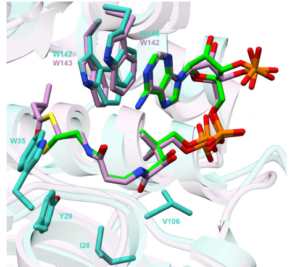
The structural basis of substrate recognition is investigated in this project. We use the tools of X-ray Crystallography (collaboration), protein-ligand docking and MD simulations to discover potent inhibitors for AHL synthases. This project will offer student training opportunities in bioinformatics, docking, homology modelling and molecular dynamics simulations.
Research Opportunities
Undergraduate Students
Please send me an email (rajnagarajan@boisestate.edu), if you are interested to participate in a research project during academic year and/or summer.
Graduate Students
If you are a prospective graduate student looking for research opportunities in my lab, please email me at rajnagarajan@boisestate.edu.
Learn more about the Masters Program in Chemistry and Biochemistry
Potential graduate students interested in applying for the interdisciplinary Ph.D program in Biomolecular Sciences, please visit Biomolecular Sciences for application information and program details.
Select Publications (2018-2023)
Patrick D. Fischer, Abu Sayeed Chowdhury, Thomas Bartholow, Shibani Basu, Eric Baggs, Huel S. Cox III, Srdan Matosin, Michael D. Burkart, Lisa Warner, Rajesh Nagarajan* and Haribabu Arthanari*. Carrier Protein Mediated Cargo Sensing in Quorum Signal Synthases, Chemical Communications (2023), 59(8), 1014-1017.
Srdan Matosin, Patrick D. Fischer, Maxim A. Droemer, Eric Baggs, Abu Sayeed Chowdhury, Isidoro Tavares, Scott B. Ficarro, Lisa Rose Warner, Haribabu Arthanari and Rajesh Nagarajan*. ₁H, ₁₃C and ₁₅N backbone and sidechain assignment of the Burkholderia mallei acyl carrier protein, J. Biomol. NMR. Assign. (2023), doi:10.1007/s12104-023-10136-4.
Shi-Hui Dong, Mila Nhu Lam, Rajesh Nagarajan and Satish K. Nair. Structure. Structure-Guided Biochemical Analysis of Quorum Signal Synthase Specificities. ACS Chemical Biology (2020), 15, 1497-1504.
Julia Thomas Oxford, Rajesh Nagarajan et al. Center of Biomedical Research Excellence in Matrix Biology: Building Research Infrastructure, Supporting Young Researchers, and Fostering Collaboration. International Journal of Molecular Sciences (2020), 21(6), 2141.
Daniel Shin, Christoph Gorgulla, Michelle E. Boursier, Neilson Rexrode, Eric C. Brown, Haribabu Arthanari, Helen E. Blackwell and Rajesh Nagarajan. N-Acyl-Homoserine Lactone Analog Modulators of the Pseudomonas aeruginosa RhlI Quorum Sensing Signal Synthase. ACS Chemical Biology (2019), 14, 10, 2305-2314.
Daniel Shin and Rajesh Nagarajan. Enzymatic Assays to Investigate Acyl Homoserine Lactone Autoinducer Synthases. Methods in Molecular Biology (2018), 1673:161-176.
Mila Nhu Lam, Dastagiri Dudekula, Bri Durham, Noah Collingwood, Eric C. Brown, and Rajesh Nagarajan. Insights into beta ketoacyl-chain recognition for beta-ketoacyl-ACP utilizing enzymes. Chemical Communications (2018), 54(64):8838-8841.
Michelle E. Boursier, Joseph D. Moore, Katherine N. Heitman, Sally P. Shepardson-Fungairino, Joshua B. Combs, Lea C. Koenig, Daniel Shin, Eric C. Brown, Rajesh Nagarajan and Helen E. Blackwell. Structure-Function Analyses of the N-Butanoyl-L-Homoserine Lactone Quorum-Sensing Signal Define Features Critical to Activity in RhlR. ACS Chemical Biology (2018), 13(9):2655-2662.
Select Grants (2019-2025)
National Science Foundation
Mechanistic Investigation on Carrier Protein Recognition in Quorum Signal Synthases
08/2019 – 07/2023
National Institutes of Health R15
Chemical Probes to Modulate Acyl-Homoserine Lactone Quorum Signal Synthesis
09/2022 – 08/2025
National Institutes of Health R21
Chemical Tools to Investigate Enzyme-Assisted Cargo-Flipping in Acyl Carrier Proteins
01/2023 – 12/2025
Teaching Philosophy
Effective classroom teaching has unique challenges in the 21 st century. Today’s students have so many career paths to choose from and often this has led to diverse interests among the student community. Even students with focused interests are faced with the daunting task of assimilating a plethora of new information. In this context, I visualize my role as a facilitator encouraging students to develop interest in the subject material and support them to understand the fundamentals of the subject instead of memorizing it. I always stress the point that earning a high grade and understanding the material goes hand-in-hand and it is often impossible to separate one from the other. I truly believe that independent thinking, not mere reiteration of facts will be critical for the success of a student in a competitive job market. As a teacher, I am committed to helping students meet these challenges and ultimately achieve their personal goals.
Teaching Roster
At Boise State (2010-present)
- CHEM111 General Chemistry I
- CHEM301: Survey of Organic Chemistry
- CHEM431: Biochemistry I
- CHEM433: Biochemistry II
- CHEM432: Biochemistry Laboratory
- BIOCHEM512: Intermediary Metabolism
- BIOCHEM513: Advanced Enzymology
- BMOL605: Current Scientific Literature
- BMOL602 Biomolecules II
At Skidmore College (2006-2010)
- CH107H: Intensive General Chemistry Honors
- CH105: Chemical Principles I
- CH105L: Chemical Principles I Laboratory
- CH106: Chemical Principles II
- CH106L: Chemical Principles II Laboratory
- CH341L: Biochemistry Laboratory
- CH342: Biochemistry II – Intermediary Metabolism
- SSP100: Drug Discovery – From Laboratory Bench to Pharmacy Stack