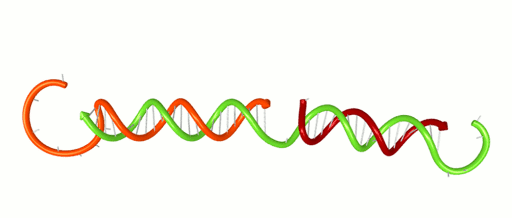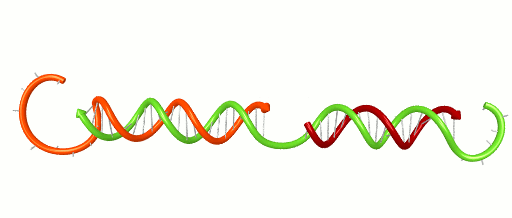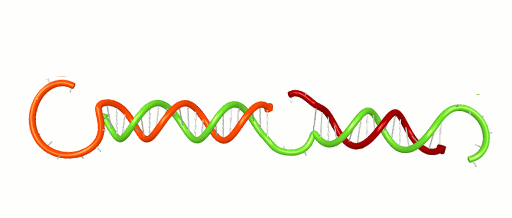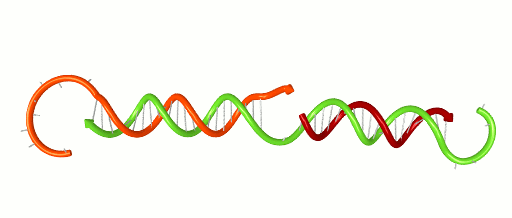
November 1, 2016 – Scientists and engineers use the rules of Watson-Crick base-pairing to design DNA systems that have the potential to perform computations and detect disease. The basic rule is that Adenine binds to Thymine and Cytosine binds to Guanine forming base-pairs through hydrogen bonding. Because hydrogen bonds are relatively weak and are on the scale of what holds water together, these bases un-pair randomly. Un-pairing is most significant at the ends of a DNA double helix (think of it as the molecule fraying like the strands at end of a rope). This fraying can have a critical impact on DNA systems. For instance, fraying can contribute to false positive signals in a DNA-based detection network. It can cause cross-talk in a DNA-based computer. Developing new design tools will help engineers avoid such problems and design better DNA systems.
Xiaoping Olson, the 1st woman to earn her PhD from the Micron School of Materials Science & Engineering, and co-workers recently reported a new design metric based on the thermodynamic properties of DNA. Analyzing their own systems, as well as those published by others, the group demonstrated the ability to locate vulnerable fraying locations and design systems with greater performance. The work, entitled “Availability: A Metric for Nucleic Acid Strand Displacement Systems,” was published recently in ACS Synthetic Biology (DOI: 10.1021/acssynbio.5b00231) and was supported in part by the W.M. Keck Foundation.