“My generation of scientists is currently trying to understand how the genome works at the molecular level,” said assistant professor Matt Ferguson. “An analogy that I like to use is that we’ve discovered a four billion year-old technology and are reverse engineering it. That’s essentially what we’re doing with the human genome, it’s like something out of a science fiction movie.”
Ferguson’s research, in collaboration with colleagues Diana Stavreva from the National Cancer Institute (NCI) of the National Institutes of Health (NIH); and Tineke Lenstra of the Netherlands Cancer Institute, Amsterdam, seeks to unravel the grammar and syntax of the human genome through a study of the regulation of transcription, or the process by with DNA is transcribed into RNA, which occurs in the nucleus of our cells. Two papers resulting from this NIH-funded research recently were published and are linked below.
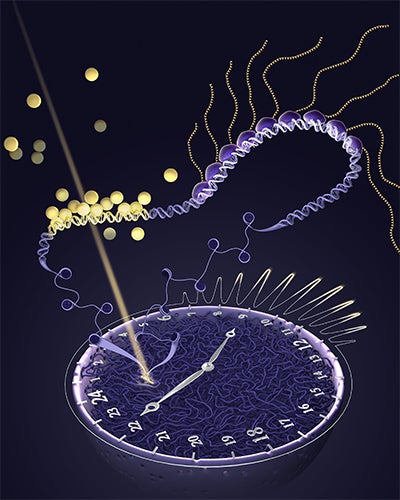
Using an innovative 3D orbital tracking microscope that Ferguson assembled in his Boise State lab, the team was able to track the sites where transcription occurs at an active gene at high spatial and temporal resolution, visualizing the process of gene activation within the nucleus of living cells for the first time.
“Nobody has ever measured the timing between the binding of a molecule, called a transcription factor, to a gene and then the resulting transcription of the RNA, so we’re the only ones that have ever made that measurement. What it can do is open a window into the living genome, as a dynamic living thing,” Ferguson said.

Gordon Hager, chief of the Laboratory of Receptor Biology and Gene Expression at NCI referred to the team’s discovery as “the holy grail” in the field. Ferguson believes the breakthrough is an essential piece to gaining a better understanding of genome biology and a new field of science that will make three things possible.
“Essentially we can fix our genomes when they are not working, curing human diseases like cancer. We can design new synthetic genomes, to do chemistry for example, and we can do DNA computation. In a sense, computation is something our genomes are already doing,” he explained.
While these three possibilities are still largely theoretical, Ferguson likens the potential for discovery and innovation with the human genome to the discoveries of early astronomers.
“Early astronomers looking at the stars knew that the stars would rise in the east, and set in the west, but there’s these wandering stars called planets. And so the most careful measurements of those wanderers led to the laws of planetary motion, which then led to developments in modern physics and modern astronomy,” he said.
For Southwestern Idaho Bridges to Baccalaureate student Iris Torres, a mechanical engineering sophomore who assists Ferguson, being a part of this breakthrough was an incredible opportunity to learn from a mentor. Through the research, she discovered an affinity and talent for research.
“When I joined the lab, I was very nervous about all the new information I had to learn, but once I started I saw how much I liked physics and biology together. I really enjoy learning more with Dr. Ferguson. He is a great mentor and the lab has helped me push myself to become better at research and science,” said Torres.
This research is funded by the Research Corporation for Science Advancement and the Gordon and Betty Moore Foundation through Grant GBMF5263.10, and by National Institute of General Medical Sciences through Grant 1R15GM123446-01.
Learn more about the team’s research below:
Stavreva DA, Garcia DA, Fettweis G, Gudla PR, Zaki G, Soni V, McGowanA, Williams G, Huynh A, Palangat M, Schiltz RL, Johnson TA, Ferguson ML, Pegoraro G, Larson DR, Upadhyaya A, Hager GL, Transcriptional bursting and co-bursting regulation by steroid hormone release pattern and transcription factor mobility. Molecular Cell. 2019.
Donovan BT, Huynh A, Ball DB, Patel H, Poirier MG, Larson DR, Ferguson ML, Lenstra TL, Live‐cell imaging reveals the interplay between transcription factors, nucleosomes, and bursting. EMBO Journal. 2019.
Advanced imaging technology reveals pulsed hormone release regulates gene transcription
https://sites.google.com/boisestate.edu/ferguson/physical-biology-of-gene-expression