DNA Nanotech
DNA Reaction Networks
From an engineering perspective, DNA is to catalytic amplifiers as silicon is to transistors. As modular components, DNA amplifiers can be interconnected to perform complex calculations chemically much the same way that transistors can be combined to perform calculations electronically. When engineered into a detection system, DNA can amplify miRNAs, perform calculations on the amplified signal, and generate an observable output, analogous to the results of a disposable pregnancy test. As miRNAs are linked to over 180 diseases, including cardiovascular, neurological, muscular, sexually transmitted, obesogenic, and diabetic diseases, the proposed research is significant because of its implications on human health. Our proposed detection system consists of four programmable DNA reaction network modules, illustrated in Fig. 1 with components color-coded by function.
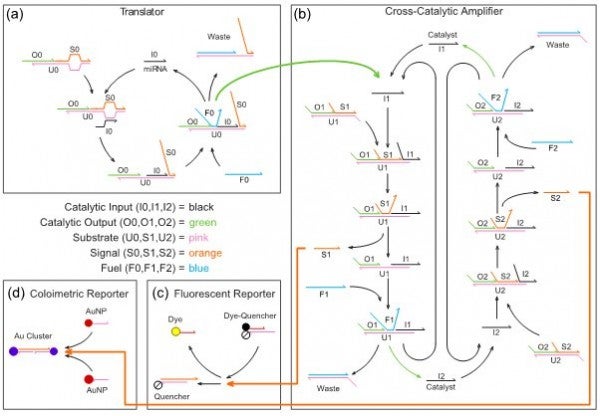
FIGURE 1. DNA reaction network engineered to detect disease-related miRNAs. Translator (a): This reaction network is engineered to detect a target miRNA. The miRNA triggers a chain reaction in which a fuel strand releases both the original miRNA and an output DNA strand. Output strands are released only if both fuel and miRNA are present. Cross-Catalytic Amplifier (b): This system of coupled reaction networks amplifies the output strand of the translator to make the miRNA presence more apparent. The translator output initiates a second chain reaction that results in exponential growth of signal strands. Fluorescent Reporter (c) and Colorimetric Reporter (d): The reporters in these two modules react with signal strands from the amplifier to produce easily observable optical signals that provide a positive/negative indication for disease.
DNA Origami for Optoelectronic Applications
The ability to precisely pattern nanoparticles is essential for realizing the potential of nanoelectronic and nanoplasmonic devices. Over the last decade, DNA oligonucleotides have been programmed to aggregate, crystallize, and self-assemble into spatially discrete assemblies and linear arrays. DNA nanotechnology offers a compelling approach towards programmable nanoparticle patterning. By implementing basic design rules, DNA can be used to form complex nanostructures using the methods of either tiled DNA motifs or DNA origami. When functionalized, these nanostructures can serve as two- and three-dimensional nanoparticle scaffolds.
Presented here is our method of fabricating nanoparticle arrays with controlled periodicity using three-dimensional, six-helix DNA origami nanotubes. DNA origami nanotubes of predetermined dimensions were used to precisely arrange nanoparticles by incorporating binding sites along the axis of the nanotube using biotin-labeled staple strands. The unique sequence of each staple strand permits precise spatial control and modular design of periodic or aperiodic binding sites. The three-dimensional DNA origami nanotubes provide a rigid structure for nanoparticle attachment in solution.
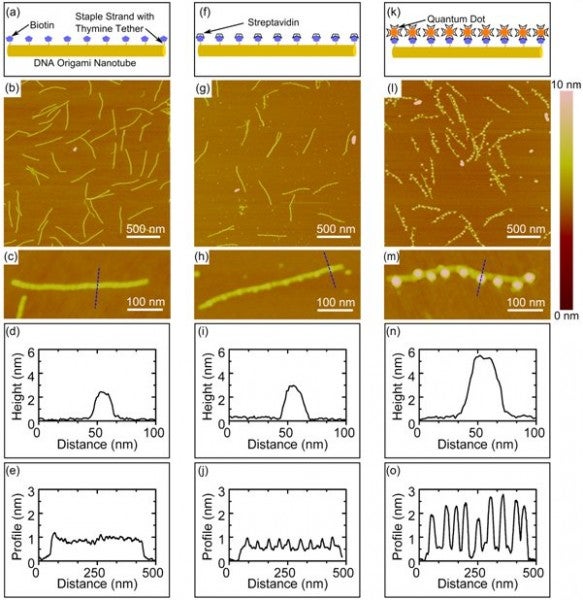
FIGURE 2. Schematics, AFM images at low magnification (upper) and high magnification (lower), and cross-sectional (upper) and axial (lower) height profiles of functionalized DNA origami nanotubes with 9 biotin binding sites with: (a-e) no attached nanoparticles; (f-j) attached streptavidin; (k-o) attached streptavidin-conjugated quantum dots. The dashed lines in the high magnification AFM images indicate the location of the cross-sectional profiles. Axial profiles represent the average of multiple profiles across the width of the nanotube.
Collaborations:
- Professor Erik Winfree – Caltech
- Dr. Paul Rothemund – Caltech
- Professor John Reif – Duke University
- Professor Dr. Fritz Simmel – Technical University of Munich
- Professor Dr. Tim Liedl – Ludwig-Maximillians University Munich (LMU) – Soft Condensed Matter Group
- Professor Cheryl Jorcyk – Boise State University
Funding:
- W.M. Keck Foundation
- DARPA
- NIH INBRE
- NIH/NCRR BRIN 2003 Seed Award Program
- *Idaho BRIN Undergraduate Summer Fellowship Program (NCRR- NIH program)
- NIH/NCRR BRIN Faculty Development Award Program (NIH/NCRR P20 RR16454)
- NSF Interdisciplinary Research (IDR) Grant